2020-01-23 00:00:00 +0000
- Janssen Vaccine start phase 1/2 clinical trials……..
- SARS-CoV (accession no. NC_004718): https://www.ncbi.nlm.nih.gov/nuccore/NC_004718.3?report=fasta
- Virus variants reflect patients severity: https://www.medrxiv.org/content/10.1101/2020.04.14.20060160v1.full.pdf+html
- 04/13/2020: xxx whole genome of COV19 were downloaded and you can download here
- 04/12/2020: Effective reproductive number (R) decrease from 3.0 to 0.3 with cordons sanitaire, traffic restriction, social distancing, home confinement, centralized quarantine, and universal symptom survey.
- 04/05/2020: The COVID-19 host genetics initiative: https://covid19hg.netlify.com/
- 04/05/2020: SITC Statement on anti-IL-6/IL-6R for COVID-19, use of IL-6 or IL-6-receptor (IL-6R) blocking antibodies like tocilizumab (Actemra, Roche-Genentech), sarilumab (Kevzara, Regeneron) and siltuximab (Sylvant, EUSA Pharma)
- 04/01/2020: How The Body Reacts To Viruses from Harvard University video
- 03/30/2020: Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2
- 03/30/2020: Janssen works together with Harvard using Janssen patented technique: AdVac and PER.C6 to develop COVID19 vaccines.
- 03/30/2020: Janssen and Biomedical Advanced Research and Development Authority (BARDA) for 1 billion $ Vaccines development
- 03/12/2020: ACE2 and TMPRSS2: The novel coronavirus 2019 (2019-nCoV) uses ACE2 and TMPRSS2 to entry target cells
- 03/06/2020:Covid-19 Small Molecule Therapies Reviewed from Science Translational Medicine
- 03/07/2020: FTF to download Virus Genome: ftp://download.big.ac.cn/Genome/Viruses/Coronaviridae/genome/
- 03/07/2020: Virus Genome Download Update: https://bigd.big.ac.cn/ncov/release_genome and https://www.gisaid.org/
- 03/03/2020: How is AI Informing the Global Health and Business Response to 2019-nCOV?
- 03/03/2020: Technical Problems with Existing CDC COVID-19 Primers, and an Improved Set of Primers.
- 02/03/2020: Dr. Zheng-Li Shi published A pneumonia outbreak associated with a new coronavirus of probable bat origin in Nature.
- 02/03/2020: Dr. Yong-Zhen Zhang lab in IBS,Fudan University published A new coronavirus associated with human respiratory disease in China in Nature
- 02/03/2020: demographics, clinical information and reason of infection were available for the first 2020 patents, click here to download
- 02/02/2020: 54 nCoV RNAs are available: https://raw.githubusercontent.com/Shicheng-Guo/2019nCoV/master/2019_nCov_genomes_2020.02.02.fasta
- 01/30/2020: Kaggle online: Daily level information on the number of 2019-nCoV affected cases across the globe:
- 01/25/2020: German Prof. Rolf Hilgenfeld, structure biologist, bring his coronavirus Substances of inhibitors to Wuhan to test the efficacy, see nature news.
- 01/24/2020: Host and infectivity prediction of Wuhan 2019 novel coronavirus using deep learning algorithm
- 01/23/2020: Discovery of a novel coronavirus associated with the recent pneumonia outbreak in 3 humans and its potential bat origin
- 01/19/2020: A mathematical model for simulating the transmission of Wuhan novel Coronavirus, see bioRxiv
- 01/24/2020: One way that viruses adapt is by encoding proteins using the same choice of codons as their host??
- 01/24/2020: primary host -> intermediate host -> human, secondary host require long time to accumulate mutation
- 01/24/2020: more discussion about 2019-nCoV in virological: http://virological.org/t/novel-2019-coronavirus-genome/319
- 01/24/2020: one-off Nextstrain build for SARS-like coronaviruses from Bedford Lab: https://github.com/blab/sars-like-cov
- 01/24/2020: ACE2 and TMPRSS2: Angiotensin I Converting Enzyme 2, functional receptor for the spike glycoprotein of the human coronaviruses SARS and HCoV-NL63.
- 01/23/2020: Phylogeny of 6 SARS-like betacoronaviruses in Wuhan.
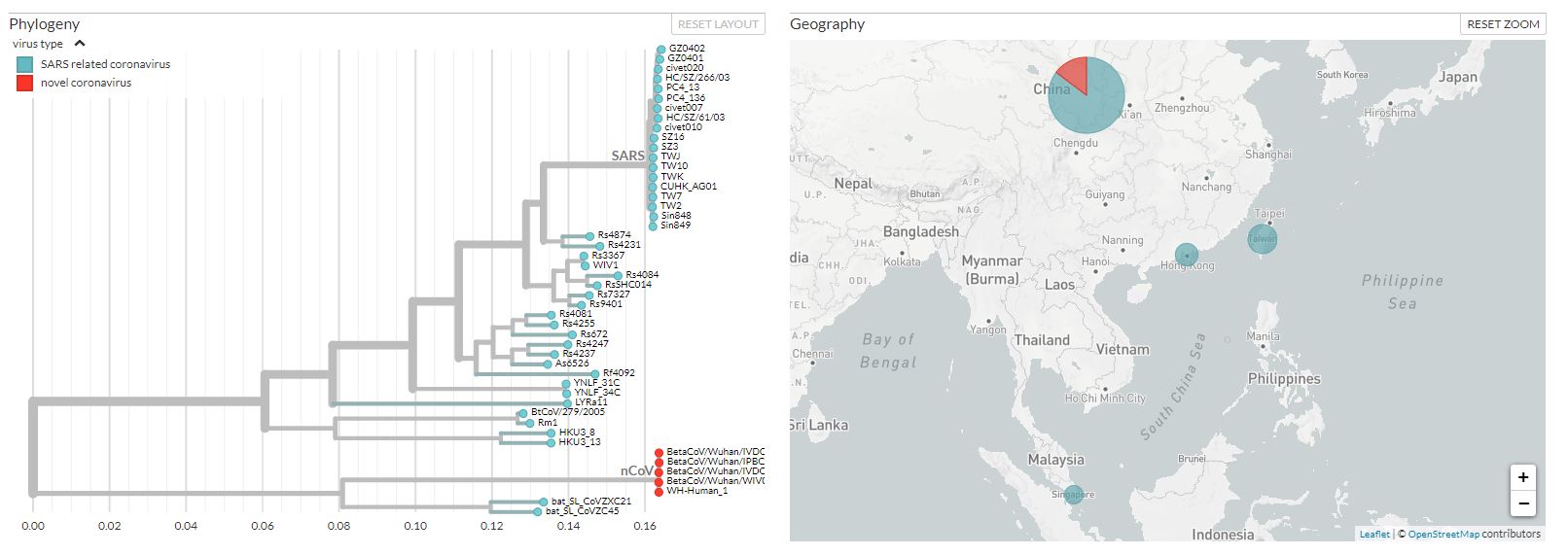
- 01/23/2020: Five new genomes have been deposited in the GISAID platform: https://gisaid.org/CoV2020
- 01/23/2020: USA confirmed the first 2019-nCoV occurred in USA in the state of Washington
- 01/23/2020: Upload fasta file to 2019nCoV and compared with other nCoV
- 01/22/2020: Dr. Shi identified bat as the virus host and upload the paper to bioRxiv
- 01/10/2020: Fudan University shared the sequence to GenBank with accession MN908947
- 12/12/2019: the first 2019-nCoV occurred in Wuhan, China and scientist think the earliest time might be 11/20/2019
2020-01-20 00:00:00 +0000
On 01/19/2020, I will give a talk about “UK Biobank Whole-Exome Sequence Binary Phenome Analysis with Robust Region-Based Rare-Variant Test, The American Journal of Human Genetics, December 19, 2019” in Marshfield Clinic Research Institute. Please check the PPT I prepared. I have implemented the pipeline in Marshfield Clinic SuperServer HPC cluster. The Figure I prepare for this post is GWAS result for colon cancer (CRC), RA and ESCC based on UKBB-50K-Exome-sequencing data. Several genes looks very interesting, for example SCL21A2.
2019-11-08 00:00:00 +0000
Aim and Background
Normal human heart tissues
Normal human blood cells
Reference
Disclosure.
- All the opinions are my own and not the views of my employer
- All the blogs are my own and not the views of my employer
- All the opinions are my own and not the views of my employer
- All the contents are my own and should never be taken seriously
- All the contents are only used for help. reminding me if misleading happens
- All the figures are only used for non-profit education. reminding me if infrigement happens
2019-10-30 00:00:00 +0000
Aim and Background
Normal human tissues
- McEWen, PNAS, 2019, The PedBE clock accurately estimates DNA methylation age in pediatric buccal cells
- Stubbs, Genome Biol, 2017,Multi-tissue DNA methylation age predictor in mouse.
- Horvath, Genome Biol, 2013, DNA methylation age of human tissues and cell types.
Cancer and adjacent tissues
Reference
Disclosure.
- All the opinions are my own and not the views of my employer
- All the blogs are my own and not the views of my employer
- All the opinions are my own and not the views of my employer
- All the contents are my own and should never be taken seriously
- All the contents are only used for help. reminding me if misleading happens
- All the figures are only used for non-profit education. reminding me if infrigement happens
2019-10-28 00:00:00 +0000
Here, I want to summarize Epigenome Research to Human Cancers with WGBS.
Reference
- Dehghanizadeh S, Khoddami V, Mosbruger TL, Hammoud SS et al. Active BRAF-V600E is the key player in generation of a sessile serrated polyp-specific DNA methylation profile. PLoS One 2018;13(3):e0192499. PMID: 29590112
- Jenkinson G, Pujadas E, Goutsias J, Feinberg AP. Potential energy landscapes identify the information-theoretic nature of the epigenome. Nat Genet 2017 May;49(5):719-729. PMID: 28346445
- Skvortsova K, Masle-Farquhar E, Luu PL, Song JZ et al. DNA Hypermethylation Encroachment at CpG Island Borders in Cancer Is Predisposed by H3K4 Monomethylation Patterns. Cancer Cell 2019 Feb 11;35(2):297-314.e8. PMID: 30753827
- Ziller MJ, Gu H, Müller F, Donaghey J et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013 Aug 22;500(7463):477-81. PMID: 23925113
- Ziller MJ, Hansen KD, Meissner A, Aryee MJ. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat Methods 2015 Mar;12(3):230-2, 1 p following 232. PMID: 25362363
Aim and Background
Disclosure
- All the opinions are my own and not the views of my employer
- All the blogs are my own and not the views of my employer
- All the opinions are my own and not the views of my employer
- All the contents are my own and should never be taken seriously
- All the contents are only used for help. reminding me if misleading happens
- All the figures are only used for non-profit education. reminding me if infrigement happens
2019-10-28 00:00:00 +0000
- McEWen, PNAS, 2019, The PedBE clock accurately estimates DNA methylation age in pediatric buccal cells
- Stubbs, Genome Biol, 2017,Multi-tissue DNA methylation age predictor in mouse.
- Horvath, Genome Biol, 2013, DNA methylation age of human tissues and cell types.
Aim and Background
Reference
Disclosure.
- All the opinions are my own and not the views of my employer
- All the blogs are my own and not the views of my employer
- All the opinions are my own and not the views of my employer
- All the contents are my own and should never be taken seriously
- All the contents are only used for help. reminding me if misleading happens
- All the figures are only used for non-profit education. reminding me if infrigement happens
2019-10-26 00:00:00 +0000
Here, I want to summarize Population Genetics in East Asian and Allele Frequency.
Aim and Background
- Z Du,2019, Genomics, Proteomics & Bioinformatics, Whole Genome Analyses of Chinese Population and De Novo Assembly of A Northern Han Genome
- S Liu,2018, Cell, Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and history in Chinese populations
- H Bai,2018, Nature Genetics, Whole-genome sequencing of 175 Mongolians uncovers population-specific genetic architecture and gene flow throughout North/East Asia
How to pre-process the plink data and association study (basic, not advanced)
cd ~/hpc/rheumatology/RA/RA500
#plink --file result_extract_forward --make-bed --out RA500
plink --bfile RA500 --mind 0.05 --make-bed --out RA2020-B1
plink --bfile RA2020-B1 --geno 0.1 --make-bed --out RA2020-B2
plink --bfile RA2020-B2 --maf 0.01 --make-bed --out RA2020-B3
plink --bfile RA2020-B3 --hwe 0.00001 --make-bed --out RA2020-B4
plink2 --bfile RA2020-B4 --king-cutoff 0.125
plink2 --bfile RA2020-B4 --remove plink2.king.cutoff.out.id --make-bed -out RA2020-B5
plink --bfile RA2020-B5 --check-sex
plink --bfile RA2020-B5 --impute-sex --make-bed --out RA2020-B6
plink --bfile RA2020-B6 --check-sex
grep PROBLEM plink.sexcheck | awk '{print $1,$2}' > sexcheck.remove
plink --bfile RA2020-B6 --remove sexcheck.remove --make-bed --out RA2020-B7
plink --bfile RA2020-B7 --test-missing midp
awk '$5<0.000001{print}' plink.missing | awk '{print $2}' > missing.imblance.remove
plink --bfile RA2020-B7 --exclude missing.imblance.remove --make-bed --out RA2020-B8
plink --bfile RA2020-B8 --pca --threads 31
# perl phen.pl RA2020-B8.fam > RA2020-B8.fam.new
# mv RA2020-B8.fam.new RA2020-B8.fam
plink --bfile RA2020-B8 --logistic --covar plink.eigenvec --covar-number 1-5 --adjust
plink --bfile RA2020-B8 --assoc --adjust gc --threads 31 --ci 0.95 --out RA500
plink --bfile RA2020-B8 --assoc mperm=1000000 --adjust gc --threads 31
Prepare Population Specific Genoytping data based on 1000 Genome Phase 3 data
#############################################
cd ~/hpc/db/hg19/beagle
for i in {1..22} X Y
do
wget http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a/b37.vcf/chr$i.1kg.phase3.v5a.vcf.gz
done
wget http://bochet.gcc.biostat.washington.edu/beagle/genetic_maps/plink.GRCh37.map.zip
wget http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a/sample_info/20140625_related_individuals.txt
wget http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a/sample_info/integrated_call_male_samples_v3.20130502.ALL.panel
wget http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a/sample_info/integrated_call_samples.20130502.ALL.ped
wget http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a/sample_info/integrated_call_samples_v3.20130502.ALL.panel
mkdir EUR
mkdir EAS
grep EUR integrated_call_samples_v3.20130502.ALL.panel | awk '{print $1}'> EUR.List.txt
grep EAS integrated_call_samples_v3.20130502.ALL.panel | awk '{print $1}' > EAS.List.txt
mkdir temp
for i in {1..22} X Y
do
echo \#PBS -N $i > $i.job
echo \#PBS -l nodes=1:ppn=1 >> $i.job
echo \#PBS -M Guo.shicheng\@marshfieldresearch.org >> $i.job
echo \#PBS -m abe >> $i.job
echo \#PBS -o $(pwd)/temp/ >>$i.job
echo \#PBS -e $(pwd)/temp/ >>$i.job
echo cd $(pwd) >> $i.job
# echo tabix -p vcf chr$i.1kg.phase3.v5a.vcf.gz >> $i.job
# echo bcftools view chr$i.1kg.phase3.v5a.vcf.gz -S EUR.List.txt -Oz -o ./EUR/chr$i.1kg.phase3.v5a.EUR.vcf.gz >>$i.job
echo bcftools view chr$i.1kg.phase3.v5a.vcf.gz -S EAS.List.txt -Oz -o ./EAS/chr$i.1kg.phase3.v5a.EAS.vcf.gz >>$i.job
qsub $i.job
done
How to prepare vcf files which can be submitted to michigan imputaiton server
cd ~/hpc/rheumatology/RA/RA500
mkdir michigan
plink --bfile RA2020-B8 --list-duplicate-vars ids-only suppress-first
plink --bfile RA2020-B8 --alleleACGT --snps-only just-acgt --exclude plink.dupvar --make-bed --out RA2020-B9
cd michigan
mkdir temp
wget https://faculty.washington.edu/browning/conform-gt/conform-gt.24May16.cee.jar -O conform-gt.24May16.cee.jar
for i in {1..23}
do
echo \#PBS -N $i > $i.job
echo \#PBS -l nodes=1:ppn=12 >> $i.job
echo \#PBS -M Guo.shicheng\@marshfieldresearch.org >> $i.job
echo \#PBS -m abe >> $i.job
echo \#PBS -o $(pwd)/temp/ >>$i.job
echo \#PBS -e $(pwd)/temp/ >>$i.job
echo cd $(pwd) >> $i.job
echo plink --bfile ../RA2020-B9 --chr $i --recode vcf-iid --out RA2020-B9.chr$i >> $i.job
echo bcftools view RA2020-B9.chr$i.vcf -Oz -o RA2020-B9.chr$i.vcf.gz >>$i.job
echo tabix -p vcf RA2020-B9.chr$i.vcf.gz >>$i.job
echo java -jar ./conform-gt.24May16.cee.jar gt=RA2020-B9.chr$i.vcf.gz match=POS chrom=$i ref=~/hpc/db/hg19/beagle/EAS/chr$i.1kg.phase3.v5a.EAS.vcf.gz out=RA2020-B9.chr$i.beagle.vcf.gz >>$i.job
echo tabix -p vcf RA2020-B9.chr$i.beagle.vcf.gz >>$i.job
qsub $i.job
done
Reference
Disclosure.
- All the opinions are my own and not the views of my employer
- All the blogs are my own and not the views of my employer
- All the opinions are my own and not the views of my employer
- All the contents are my own and should never be taken seriously
- All the contents are only used for help. reminding me if misleading happens
- All the figures are only used for non-profit education. reminding me if infrigement happens
2019-10-24 00:00:00 +0000
Recently, I need to give a talk about genetic and epigenetic based Intra-Tumor Heterogeneity (ITH)
Aim and Background
Reference
- Jones, H.G. et al. Genetic and Epigenetic Intra-tumour Heterogeneity in Colorectal Cancer. World J Surg 41, 1375-1383 (2017).
Disclosure.
- All the opinions are my own and not the views of my employer
- All the blogs are my own and not the views of my employer
- All the opinions are my own and not the views of my employer
- All the contents are my own and should never be taken seriously
- All the contents are only used for help. reminding me if misleading happens
- All the figures are only used for non-profit education. reminding me if infrigement happens
2019-10-12 00:00:00 +0000
Recently, I need to give a talk about electronic health/medical records (EHR/EMR) data analysis. Why we need analyze EHR and EMR data? What’s the benefit and what’s the challenge? How to apply text-mining and data-mining in non-structural or semi-structual data analysis? How to deal with compliance and HIPPA? How to deal with multiple EMR systems and diffential format? the importance of genetic/genomic information in health care managment? the importance of the heatlh history of the patients? the mature pipeline to extract important clinical information? cancer real-world data (RWD) analysis? NCBI pubmed data re-leanring? How to connect bioinformatics team and data science as well as medical informatic scientist?
Aim and Background
Benefit of Predictive analytics based on complete and relevant EMR data:
- More accurate diagnoses
- Improved public health and preventive medicine
- Decreased healthcare costs
Challenge of electronic health/medical records (EHR/EMR) data analysis
Reference
- https://archer-soft.com/en/blog/importance-healthcare-data-analytics-emr
Disclosure.
- All the opinions are my own and not the views of my employer
- All the blogs are my own and not the views of my employer
- All the opinions are my own and not the views of my employer
- All the contents are my own and should never be taken seriously
- All the contents are only used for help. reminding me if misleading happens
- All the figures are only used for non-profit education. reminding me if infrigement happens
2019-10-08 00:00:00 +0000
How to apply Artificial Intelligence in medical diagnosis and medical device from US-FDA policy? More and more patents based on artifical intelligence occured in medical diagnosis and medical service, however, FDA have its own policy in the usage of AI in medical production. Here, we discussed the best approach to design machine learning and AI in production strategy design. Usually, we will have training dataset and test dataset in the machine learning modeling. Here, I will use an example to introduce the best way to apply AI in your production design.
Footnote
- All the opinions are my own and not the views of my employer
- All the blogs are my own and not the views of my employer
- All the opinions are my own and not the views of my employer
- All the contents are my own and should never be taken seriously
- All the contents are only used for help. reminding me if misleading happens
- All the figures are only used for non-profit education. reminding me if infrigement happens

